Color key to variant forms
This key was last updated June 30, 2021.
In the cov.lanl.gov website we have several tools where track common variants of Spike, both over time and geographically. Not all of these lineages are currently expanding, but all were a significant part of a regional sampling at some point in 2021. We note in the key the Pangolin lineage designation associated with these variant forms of the Spikes we are tracking, but please note that our web site tracks the actual most common Spike found in the lineage, plus variants with additional mutations in that backbone, but not the Pangolin lineage itself. We do not include sequences that are likely ancestral forms of these virus, with partial sets of the listed amino acids, in our tracking summaries. The Greek letter WHO designation for these variants is also noted.
A few of the simpler forms we are tracking requires an exact match: the G clade with just G614D, and several other Spike variants with just single mutations in addition to G614D. This is to distinguish these single Spike mutations from expanding lineages which include those specific mutations.
Please note our input data is a filtered set of GISAID data, that excludes incomplete or problematic sequences, as described in Korber et al. (Cell 182:4 812-827).
NOTE: We are NOT tracking insertions in Spike sequences in this output; insertions are still very rare, but are found on occasion.
In particular, we have found them associated with a few rare Pango lineages including:
| B.1.621 | T95I, insert144T, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, D950N |
| A.2.5.2 | del141-143, insert215AGG, D215Y, L452R, D614G |
| AT.1 | P9L, del136-144, D215G, H245P, E484K, D614G, N679K, insert679GIAL, E780K |
| B.1.214.2 | insert214TDR, Q414K, N450K, D614G, T716I |
Where 'insert' indicates an insertion at the given position followed by the list amino acids added, and 'del' indicates a deletion.
Color key view 1 PDF
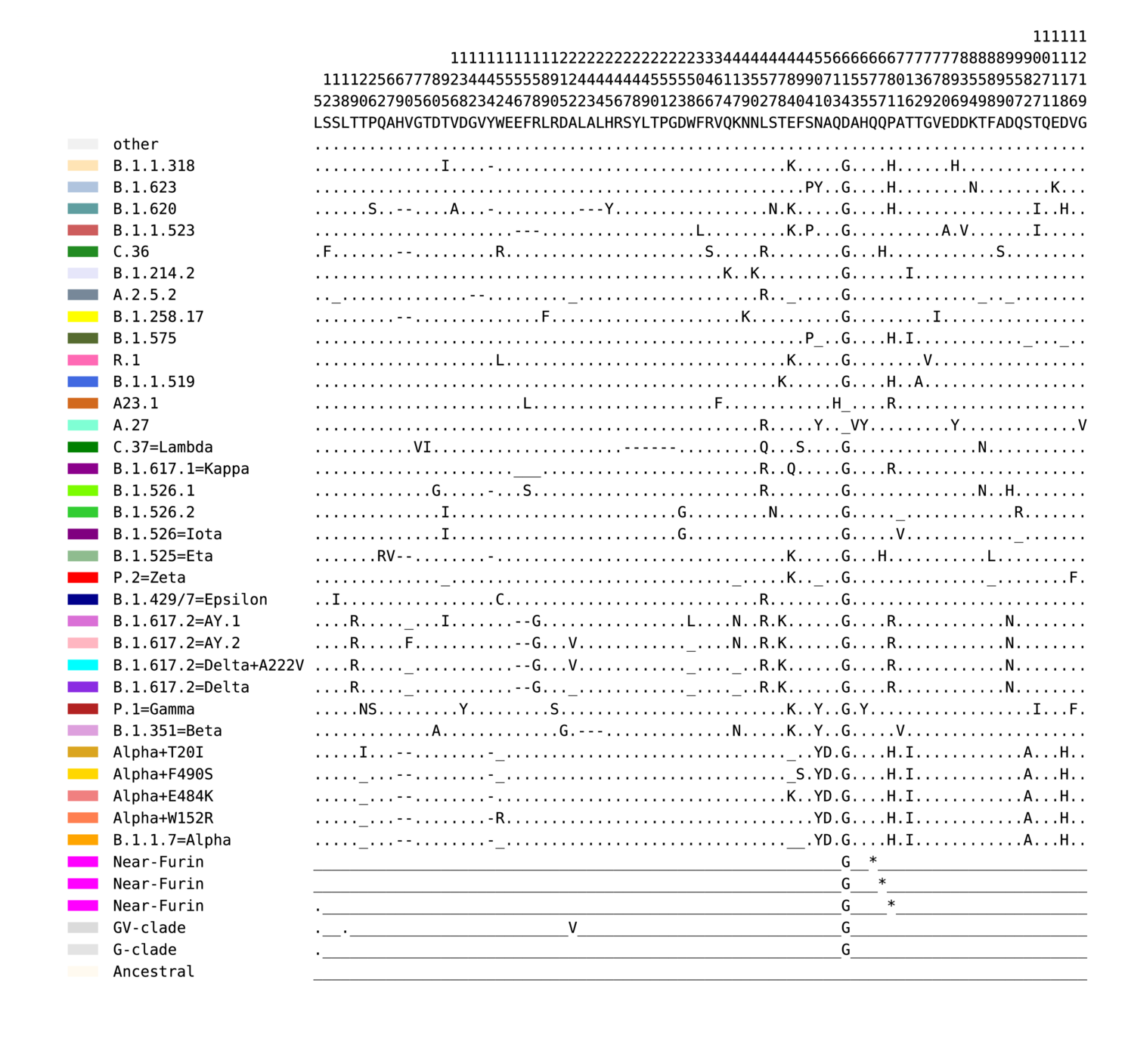
In view 1, a period (.) indicates any amino acid allowed in this position, a dash (-) is a deletion, and an underscore (_) specifically requires the ancestral amino acid. Positions numbers are written vertically, and all positions required to differentiate between common circulating forms of spike are included in the mini-alignment. In some cases, for example the Delta variant, there are two very common forms at two key positions T95I/T and G142D/G. Here we enable either form to be grouped with Delta, to include these common variants in our summaries; the actual most common form includes the mutations T95I and G142D, but a virus with the ancestral state in these positions will also be included in our tracking.
In view 2, the (-) represents a deletion.
Color key view 2 PDF

View 3 shows the full mutation string for the most common sequence that matches the pattern.
Color key view 3 PDF

Questions or comments? Contact us at seq-info@lanl.gov.




