SHIVER
SARS CoV-2 Historically Identified Variants in Epitope Regions
Variants: Delta and Omicron Variants
Strategy: Take turns
Reference strain: Wuhan reference
SHIVER: SARS CoV-2 Historically Identified Variants in Epitope Regions
SHIVER identifies variant forms of the SARS CoV-2 virus with a focus on the NTD and RBD neutralizing antibody epitope regions of the Spike protein, as well as sites related to furin cleavage; the forms are chosen to maximize coverage globally and/or on separate continents[*], depending on which of several strategies is employed.
In the Table of Variants, below, the first column is the pattern at sites where differences occur, relative to initial (Wuhan) sequence, with site numbers read down vertically.
Table of Variants
LPM = Local Pattern Matches = # of seqs in continent that match over epitope region
GPM = Global Pattern Matches = # of seqs in world that match over epitope region
GSM = Global Sequence Matches = # of seqs in world that match over whole Spike protein
111122222333333333334444444444444444444444455666 11111445545556333455677770001344445555677788889900578 66669467854684259668713563587504560256057813463815591 Name LPM GPM GSM GSM/GPM [Mutations] (Lineage) ----TYHFRHSSWAILGRKIVSSSTRDRKANKVGNLLFNASTNVEFQQNYHNP 1-Initial 0 0 0 0.0% [] MPLFI-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 2-North-America-1 4587 6820 3309 48.5% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] ....I-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 3-Europe-1 108 570 289 50.7% [T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 4-Asia-1 63 791 121 15.3% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,Q183H,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.K.HSDWSVK.N.K-KP.RYHYKR 5-Oceania-1 5 82 56 68.3% [+16MPLF,T19I,R21T,T22N,L24-,P25-,P26-,A27S,S31F,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,K182N,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456V,N460K,S477N,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DV.HTT.AFPF.KNSN.K.HSDWSLK.NKK-KP.RYHYKR 6-United-Kingdom-1 1 2 1 50.0% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,F32-,S50L,W64R,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,V367A,S371F,S373P,S375F,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,T761I,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-QSGN...DV.HTT..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 7-South-America-1 9 329 179 54.4% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,H146Q,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 8-Africa-1 1 57 28 49.1% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] ....I-.SGN...DV.HTT..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 9-North-America-2 107 163 42 25.8% [T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,Q183H,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.K.PSDWSLK.NKK-KP.RYHYKR 10-Europe-2 68 164 115 70.1% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,Q183H,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445P,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] ....I-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 11-Asia-2 47 47 19 40.4% [T19I,R21T,L24-,P25-,P26-,A27S,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DVSHTT..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 12-Oceania-2 2 13 5 38.5% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,Q183H,N211-,L212I,V213G,L216F,H245N,A264D,I332V,L335S,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN..RDV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 13-United-Kingdom-2 1 4 3 75.0% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,W258R,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] ....I-QSGN...DV.HTT..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 14-South-America-2 2 49 35 71.4% [T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,H146Q,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNSNSK.HSDWSLK.NKK-KPERYHYKR 15-North-America-3 87 133 63 47.4% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,A435S,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKN.N.K.HSDWSLK.NKK-KPERYHYKR 16-Europe-3 23 66 18 27.3% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KP.RYHYKR 17-Asia-3 45 63 23 36.5% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGNF..DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 18-Oceania-3 2 38 30 78.9% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,S254F,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.HST..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 19-United-Kingdom-3 1 1 1 100.0% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346S,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] ....I-.SGN...DV.H.T..FPFAKN.N.K.HSDWSLK.NKK-KPERYHYK- 20-South-America-3 1 1 1 100.0% [T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681-,A684X,R685X,S686X,V687X,A688X,S689X,Q690X,S691X,I692X,I693X,A694X,Y695X,T696X,M697X,S698X,L699X,G700X,A701X,E702X,N703X,S704X,V705X,A706X,Y707X,S708X,N709X,N710X,S711X,I712X,A713X,I714X,P715X,T716X,N717X,F718X,T719X,I720X,S721X,V722X,T723X,T724X,E725X,I726X,L727X,P728X,V729X,S730X,M731X,T732X,K733X,T734X,S735X,V736X,D737X,C738X,T739X,M740X,Y741X,I742X,C743X,G744X,D745X,S746X,T747X,E748X,C749X,S750X,N751X,L752X,L753X,L754X,Q755X,Y756X,G757X,S758X,F759X,C760X,T761X,Q762X,L763X,N764X,R765X,A766X,L767X,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYK- 21-North-America-4 36 47 3 6.4% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681-,A684X,R685X,S686X,V687X,A688X,S689X,+689X,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNS..K.HSDWSLK.NKK-KPERYHYKR 22-Europe-4 22 22 9 40.9% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.KRHSDWSLKVNKK-KP.RYHYKR 23-Asia-4 23 28 22 78.6% [+16MPLF,T19I,R21T,T22N,L24-,P25-,P26-,A27S,S31P,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,K182R,R190S,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,K444R,V445H,G446S,N450D,L452W,L455S,F456L,N460K,A475V,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.KRHSDWSLK.NKK-KP.RYHYKR 24-Oceania-4 2 40 27 67.5% [+16MPLF,T19I,R21T,T22N,L24-,P25-,P26-,A27S,S31P,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,K182R,R190S,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,K444R,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYRR 25-United-Kingdom-4 1 14 2 14.3% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679R,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] ....I-.SGN...DV.H.TV.FPFAKN.N.K.HSDWSLK.NKK-KPERYHYK- 26-South-America-4 1 1 1 100.0% [T19I,R21T,L24-,P25-,P26-,A27S,S31-,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,I358V,S371F,S373P,S375F,T376A,R403K,D405N,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,T572I,D614G,P621S,H655Y,N679K,P681-,A684X,R685X,S686X,V687X,A688X,S689X,Q690X,S691X,I692X,I693X,A694X,Y695X,T696X,M697X,S698X,L699X,G700X,A701X,E702X,N703X,S704X,V705X,A706X,Y707X,S708X,N709X,N710X,S711X,I712X,A713X,I714X,P715X,T716X,N717X,F718X,T719X,I720X,S721X,V722X,T723X,T724X,E725X,I726X,L727X,P728X,V729X,S730X,M731X,T732X,K733X,T734X,S735X,V736X,D737X,C738X,T739X,M740X,Y741X,I742X,C743X,G744X,D745X,S746X,T747X,E748X,C749X,S750X,N751X,L752X,L753X,L754X,Q755X,Y756X,G757X,S758X,F759X,C760X,T761X,Q762X,L763X,N764X,R765X,A766X,L767X,T768X,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.HTT..FPFAKNSN.K.HSDWSVK.NKK-KP.RYHYKR 27-North-America-5 33 55 19 34.5% [+16MPLF,T19I,R21T,T22N,L24-,P25-,P26-,A27S,S31F,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,K182N,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,R346T,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456V,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L] MPLFI-.SGN.L.DV.H.T..FPFAKNSN.K.HSDWSLK.NKK-KPERYHYKR 28-Europe-5 15 18 10 55.6% [+16MPLF,T19I,R21T,T22N,L24-,P25-,P26-,A27S,S50L,F59S,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,N211-,L212I,V213G,L216F,H245N,S256L,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478K,N481K,V483-,E484K,F486P,Q493E,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,V1104L,P1143L] MPLFI-.SGN...DV.H.T..FPFAKNSN.K.HSDWSLK.NIK-KP.RYHYKR 29-Asia-5 14 17 4 23.5% [+16MPLF,T19I,R21T,L24-,P25-,P26-,A27S,S50L,H69-,V70-,V127F,G142D,Y144-,F157S,R158G,G184S,N211-,L212I,V213G,L216F,H245N,A264D,I332V,G339H,K356T,S371F,S373P,S375F,T376A,R403K,D405N,R408S,K417N,N440K,V445H,G446S,N450D,L452W,L455S,F456L,N460K,S477N,T478I,N481K,V483-,E484K,F486P,Q498R,N501Y,Y505H,E554K,A570V,D614G,P621S,H655Y,N679K,P681R,N764K,D796Y,S939F,Q954H,N969K,P1143L]
In the Table of Variants, above, the first variant in the input alignment is taken as the reference sequence, and is the ancestral Wuhan variant to ensure epitope regions are chosen appropriately. The alignment on the left shows the positions that define unique common forms that are searched using SHIVER. The positions numbers are written vertically. The amino acids in the top row are taken from is the ancestral Wuhan variant. The epitope regions in Spike that are explored for a focused search for the common Spike variants are defined at the end of this document. The epitope and furin cleavage regions in Spike that are featured are defined below.
The basic NTD supersite sites selected are for inclusion are based on:
Sites 14-20, 140-158, and 245-264: McCallum, M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:9 2332-2347.e16 (2021)
Site 13: Impacts signal peptide cleavage and NTDss antibodies. McCallum, M. et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science 373:648-654 (2021)
Sites 242-244: Impacts NTDss antibody potency SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma Wibmer, C. et al. Nature Med. 27(4): 622-625.
Toggling Sites: Site 18 is in the NTDss and toggles frequently between L and F, so we exclude it from the tallies of forms of the regions of interest as it splits the counts on otherwise distinctive forms. An analogous situation is a problem for site 142. Among Delta variants, every common variant within the Delta lineages includes both (the ancestral) G and D at site 142. This is because the ARTIC 3 primers can results in an erroneous call of the ancestral G at position 142. The G142D mutation is the common form, and this error is resolved by using the ARTIC 4 primers. By excluding both sites 18 and 142 from our NTDss definition, we group the forms of Spike that carry either form in our tallies.
Analysis of the ARTIC version 3 and version 4 SARS-CoV-2 primers and their impact on the detection of the G142D amino acid substitution in the spike protein. Davies et al. bioRxiv 10.1101/2021.09.27.461949 (2021)
Sites 330-521: the RBD region includes positions 330-521, based on a synthesis of the literature from early 2020.
Furin related sites: mutations that add positive charge to near the furin cleavage site can enhance Spike cleavage and infectivity. Also, the change at H655Y (Alba2021) has been shown to impact furin cleavage, and we include site 950 as it accompanies P681R in Delta and P681H in Mu, to variants that were particularly fast spreading, though Delta became prevalent. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. Saito et al. bioRxiv 10.1101/2021.06.17.448820 (2021) SARS-CoV-2 variants of concern have acquired mutations associated with an increased spike cleavage. Alba et al. bioRxiv 10.1101/2021.08.05.455290 (2021)
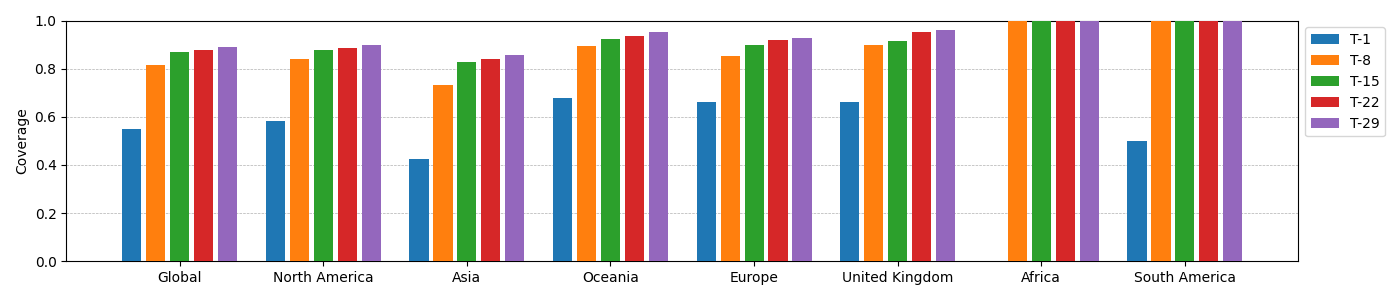
Table of Coverages
In table below, T-n refers to a batch of the first n variants. Coverage is defined as fraction of sequences in the continent with an exact match (over the region NTDss-18-142+RBD+furin) to one of the first n variants. (Here, 'T' corresponds to the 'Taketurns' strategy.) The coverage table is based on 10884 sequences.
Continent Name Coverage
Global T-1 0.0000
North-America T-1 0.0000
Europe-minus-United-Kingdom T-1 0.0000
Asia T-1 0.0000
Oceania T-1 0.0000
United-Kingdom T-1 0.0000
South-America T-1 0.0000
Africa T-1 0.0000
Global T-8 0.7948
North-America T-8 0.8177
Europe-minus-United-Kingdom T-8 0.8066
Asia T-8 0.5733
Oceania T-8 0.8361
United-Kingdom T-8 0.8548
South-America T-8 0.7179
Africa T-8 1.0000
Global T-15 0.8475
North-America T-15 0.8635
Europe-minus-United-Kingdom T-15 0.8539
Asia T-15 0.6950
Oceania T-15 0.8730
United-Kingdom T-15 0.9032
South-America T-15 0.8462
Africa T-15 1.0000
Global T-22 0.8693
North-America T-22 0.8805
Europe-minus-United-Kingdom T-22 0.8788
Asia T-22 0.7478
Oceania T-22 0.8934
United-Kingdom T-22 0.9194
South-America T-22 0.8718
Africa T-22 1.0000
Global T-29 0.8852
North-America T-29 0.8893
Europe-minus-United-Kingdom T-29 0.8990
Asia T-29 0.8082
Oceania T-29 0.9098
United-Kingdom T-29 0.9355
South-America T-29 0.8974
Africa T-29 1.0000
This run uses the T=taketurns strategy for identifying further variants. Each continent, in turn, chooses the next variant, based on which is the most common variant in that continent that has not already been chosen. The order of the continents is based on number of samples available in those continents.
Sequence sample dates range from 2024-09-14 to 2024-10-14. The number of sequences, broken out by continent is: Total: 10884, North-America: 7280, Europe-minus-United-Kingdom: 2327, Asia: 928, Oceania: 244, United-Kingdom: 62, South-America: 39, Africa: 4. The focus here is specifically on the epitope region: NTDss-18-142+RBD+furin Sites: 13-17,19,20,140,141,143-158,242-264,330-521,655,675,677,679,681,950
[*] Note that the UK is treated as a separate continent because so much of the sequencing has been from the UK.
Questions or comments? Contact us at seq-info@lanl.gov.




