This website provides analyses and tools for exploring accruing mutations in hCoV-19 (SARS-CoV-2) geographically and over time, with an emphasis on the Spike protein, using data from GISAID.
The SARS-CoV-2 sequence data used for these analyses was updated from GISAID on Mar 6, 2026.
With the ever growing database of sequences in GISAID, sometimes the web connection times out before the analysis is complete. If you have this problem, please check "email results" and an email with a link will be sent to you when the job is complete.
Frequencies of Key Pango Lineage Groupings Over Time
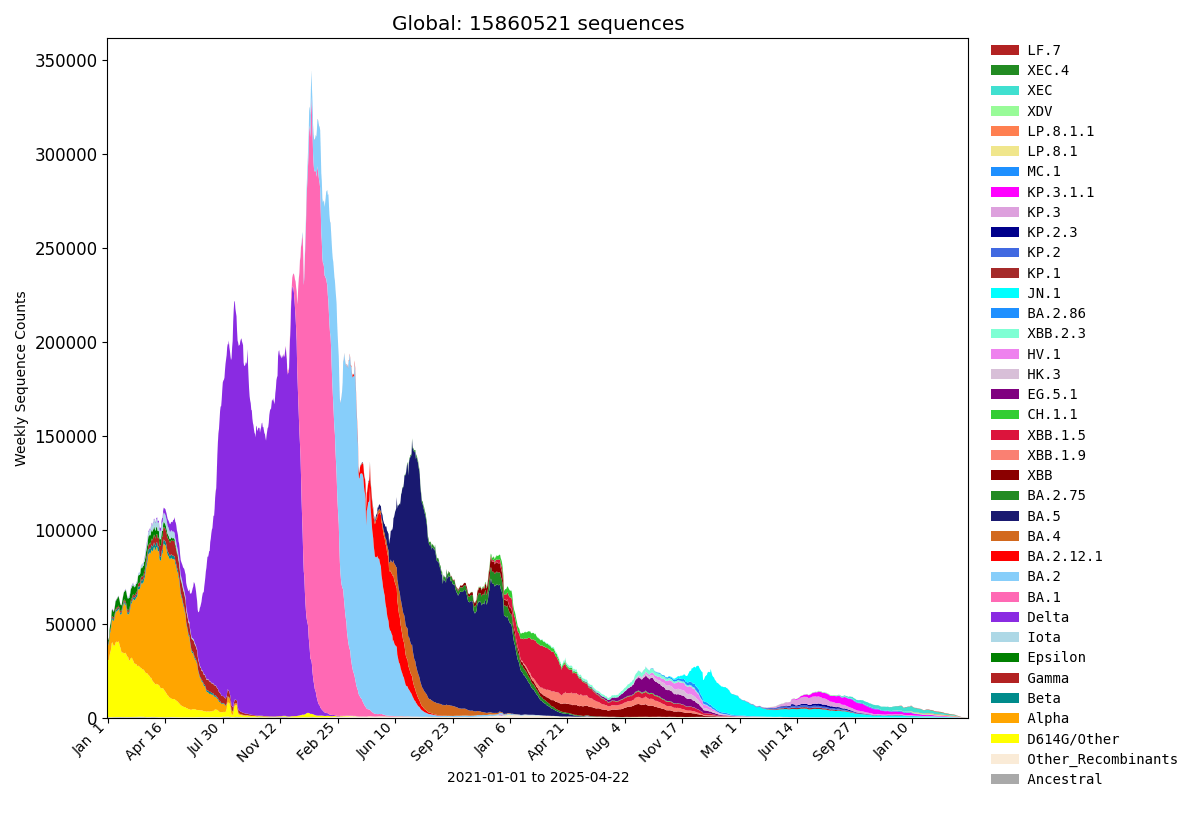
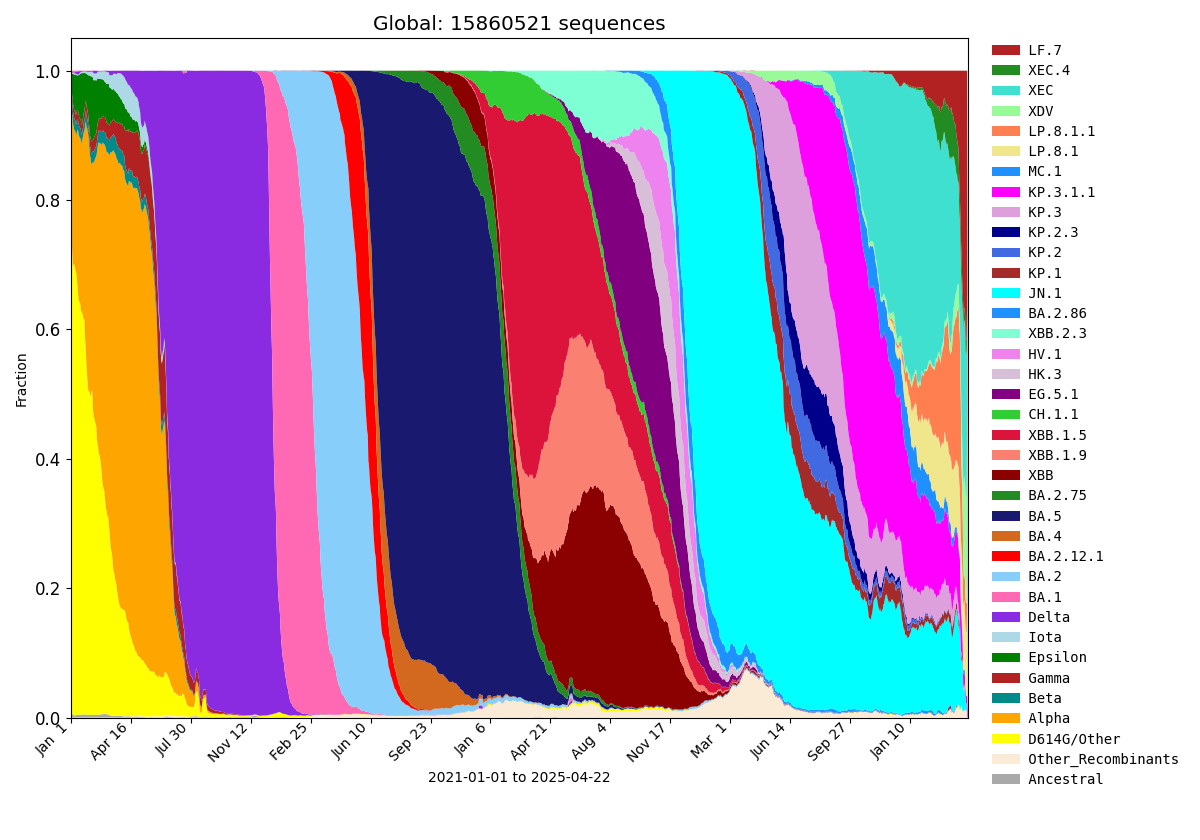
Data Summaries
These analyses are updated several times a week. Our web-based tools can focus on specific geographic regions and time frames, using the links listed under “Tools” in the blue menu bar.
| Name | Description |
|---|---|
| SARS-CoV-2 map |
Geographical summaries of Spike variants displayed as pie charts on a map |
| Embers | Transitions in common Spike variants over time globally and by continent |
| Mutating Sites in Spike | Table summarizing the amino acid variation and entropy at each site in Spike |
| SHIVER | Highlights recently emergent Spike epitope variants globally and by continent |
| Xspike | Highlights recent forms of Spike carrying the most variable amino acids and covariation patterns |
| Pango Lineage Spike Variants | Tallies of Spike variant forms assigned to each Pango Lineage |
| Spike Variants | Listing of representative forms of Spike variants of current or historical interest |
Current data shared through GISAID
How to cite
Acknowledgements
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID on which this research is based. The original data are available from https://www.gisaid.org
This COVID-19 response analysis pipeline is supported by: The Laboratory Directed Research and Development program of Los Alamos National Laboratory (20200706ER), and by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Interagency Agreement No. AAI12007-001-00000.
GISAID data provided on this website is subject to GISAID’s Terms and Conditions




