Embers
Embers: follow emergent SARS-CoV-2 variants over time
Run Geographically Restricted Embers
Last update: May 10, 2024
Delta and Omicron Variants
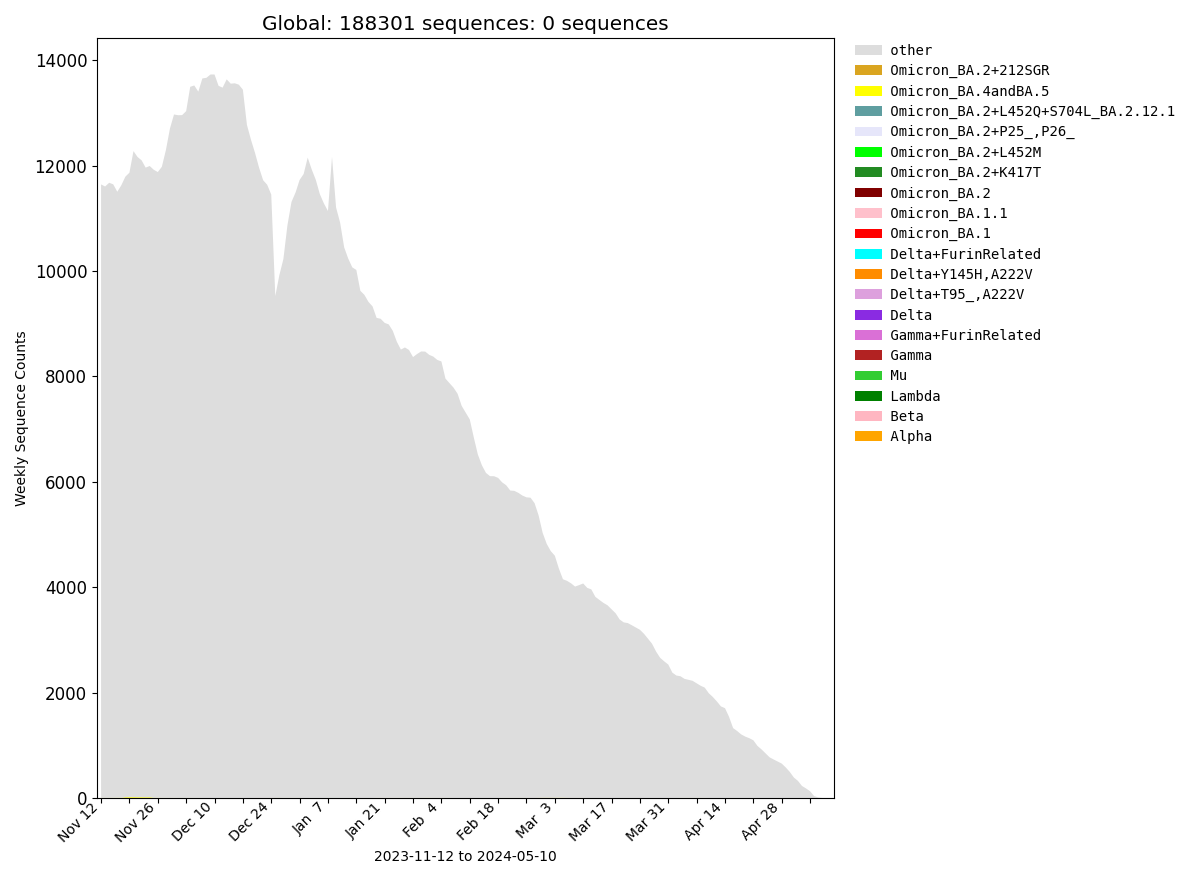
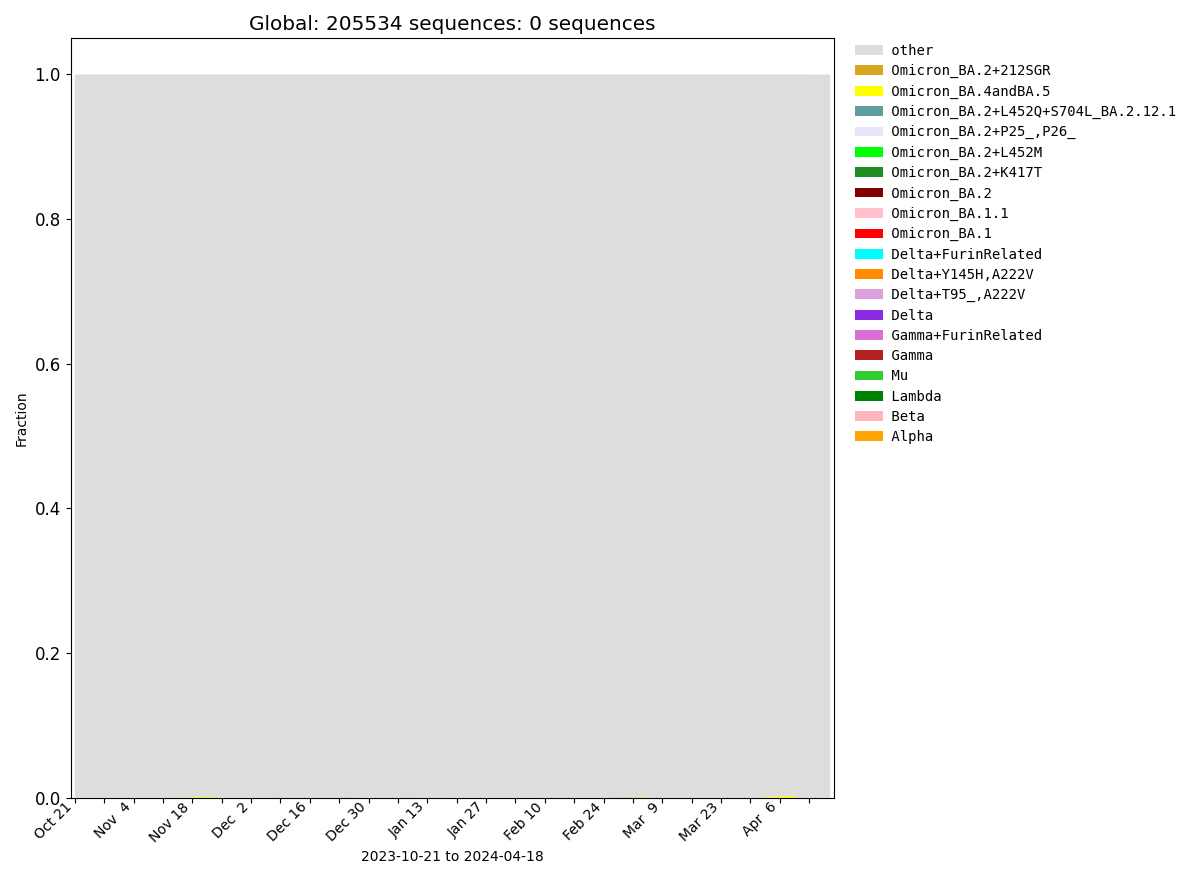
Global
| Frequencies of Key Pango Lineage Groupings Over Time |
 |
 |
Variants and Counts
1111
1111111111222222222222222222222222223333334444444444444455556666667777788999990112
1122222266777893444455559111111111112444444455555893477770014457788999900471577780016955556882176
8904567979056058234526780122223444452234678901238999613565870627846036815704557911464669049127864
LTTLPPATAHVGTDTDGVYYWEFRRNL---VR---DALALRSYLTPGDWVTGRSSSTDRKNGLSTEFFQGQNYTADHQQNPASTNDNTDQNLSTDVV Counts Percent Lineage
.I..............D..........SGRG.---................D.FPFANSNK._NKA..R.RYH..GY..KH...KY...HK...... 0 0.00% Omicron_BA.2+212SGR
.I.........................---G.---................D.FPFANSNK.RNKAV._.RYH..GY..KH...KY...HK...... 29 0.02% Omicron_BA.4andBA.5
.I..............D..........---G.---................D.FPFANSNK.QNKA..R.RYH..GY..KH.L.KY...HK...... 1 0.00% Omicron_BA.2+L452Q+S704L_BA.2.12.1
.I..__..........D..........---G.---................D.FPFANSNK._NKA..R.RYH..GY..KH...KY...HK...... 0 0.00% Omicron_BA.2+P25_,P26_
.I..--..........D..........---G.---................D.FPFANSN..MNKA..R.RYH..GY..KH...KY...HK...... 0 0.00% Omicron_BA.2+L452M
.I..............D..........---G.---................D.FPFANSTK..NKA..R.RYH..GY..KH...KY...HK...... 2 0.00% Omicron_BA.2+K417T
.I..--..........D..........---G.---................D.FPFANSN.._NKA..R.RYH..GY..KH...KY...HK...... 2 0.00% Omicron_BA.2
........V--...I............---..---................DKLPF.........A..R.RYH..GY..KH...KY...HK...... 1 0.00% Omicron_BA.1.1
........V--...I............---..---................D_.PF___..._..A..R.RYHK.GY..KH...KYK..HKF..... 0 0.00% Omicron_BA.1
........V--...I............---..EPE................D_.PF......_..A..R.RYH..GY..KH...KY...HK...... 0 0.00% Omicron_BA.1
.R........_..........--G...---..---............._.........._..R.K..........G...*........N........ 0 0.00% Delta+FurinRelated
.R........_..........--G...---..---............._.........._..R.K..........G..*.........N........ 0 0.00% Delta+FurinRelated
.R........_..........--G...---..---............._.........._..R.K..........G.*..........N........ 0 0.00% Delta+FurinRelated
.R........_..........--G...---..---............._.........._..R.K..........G....H.......N........ 0 0.00% Delta+FurinRelated
.R........_..........--G...---..---............._.........._..R.K..........GY...........N........ 0 0.00% Delta+FurinRelated
.R........_..........G--...---..---............._.........._..R.K..........G...*........N........ 0 0.00% Delta+FurinRelated
.R........_..........G--...---..---............._.........._..R.K..........G..*.........N........ 0 0.00% Delta+FurinRelated
.R........_..........G--...---..---............._.........._..R.K..........G.*..........N........ 0 0.00% Delta+FurinRelated
.R........_..........G--...---..---............._.........._..R.K..........G....H.......N........ 0 0.00% Delta+FurinRelated
.R........_..........G--...---..---............._.........._..R.K..........GY...........N........ 0 0.00% Delta+FurinRelated
.R........_...I....H.G--...---..---.V..........._.........._..R.K..........G____R.......N........ 0 0.00% Delta+Y145H,A222V
.R........_...I....H.--G...---..---.V..........._.........._..R.K..........G____R.......N........ 0 0.00% Delta+Y145H,A222V
.R........_..._......G--...---..---.V...........__........._..R.K..........G____R.......N......._ 0 0.00% Delta+T95_,A222V
.R........_..._......--G...---..---.V...........__........._..R.K..........G____R.......N......._ 0 0.00% Delta+T95_,A222V
.R....._.._..........G--...---..---._.......__.._._........_..R.K_.........G____R.......N......._ 0 0.00% Delta
.R....._.._..........--G...---..---._.......__.._._........_..R.K_.........G____R.......N......._ 0 0.00% Delta
........................S..---..---..............................K.....Y...GY...*..............F. 0 0.00% Gamma+FurinRelated
........................S..---..---..............................K.....Y...GY..*...............F. 0 0.00% Gamma+FurinRelated
........................S..---..---..............................K.....Y...GY.*................F. 0 0.00% Gamma+FurinRelated
........................S..---..---..............................K.....Y...GY*.................F. 0 0.00% Gamma+FurinRelated
........................S..---..---..............................K.....Y...GY____..............F. 0 0.00% Gamma
..............I...SN.......---..---.................K............K.....Y...G....H.......N........ 0 0.00% Mu
...........VI..............---..---......-------..............Q....S.......G...........N......... 0 0.00% Lambda
.............A.............---..---G.---...................N.....K.....Y...G.....V............... 0 0.00% Beta
.._......--.......-._......---..---....................................Y..DG....H.._........A.H.. 0 0.00% Alpha
.._......--.......-._......---..---....................................Y..DG....R..I........A.H.. 0 0.00% Alpha
.._......--......._._......---..---....................................Y..DG....H..I........A.H.. 0 0.00% Alpha
.._......--.......-._......---..---....................................Y..DG....H..I........A.H.. 1 0.00% Alpha
The most common sequence patterns that are missed by the varint patterns
Missing: 3375 patterns, for a total of 188265 sequences
1111
1111111111222222222222222222222222223333334444444444444455556666667777788999990112
1122222266777893444455559111111111112444444455555893477770014457788999900471577780016955556882176
8904567979056058234526780122223444452234678901238999613565870627846036815704557911464669049127864
LTTLPPATAHVGTDTDGVYYWEFRRNL---VR---DALALRSYLTPGDWVTGRSSSTDRKNGLSTEFFQGQNYTADHQQNPASTNDNTDQNLSTDVV Counts Percent NearbyLineage(s)
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 95683 50.81%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 23067 12.25%
_I_---S_________D_-___L____---E_---___________V____HTFPFANSNKSRNKAPS__RYH__GY__KH___KY___HK______ 15586 8.28%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NRAPS__RYH__GY__KH___KY___HK______ 6347 3.37%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NKAPS__RYH__GY__KH_L_KY___HK______ 6208 3.30%
_I_---S__--_____D_-___SG_-I---G_---________________HTFPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 4099 2.18%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NRAPS__RYH__GY__KHV__KY___HK______ 2664 1.41%
_I_---S__--_____D_-___SG___---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 1574 0.84%
_I_---S_________D_-________---E_---____________G___HTFPFANSNKS_NRAPS__RYH__GY__KH___KY___HK______ 1513 0.80%
_I_---S_________D_-___L____---E_---___________V____HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 1497 0.80%
_I_---S_________D_-________---E_---_______H___V____HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 1271 0.67%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFAN_NKSWNKKP___RYH_VGY__KR___KY___HK______ 933 0.50%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANS_KSWNKKP___RYH_VGY__KR___KY___HK______ 695 0.37%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NKAPP__RYH__GY__KRV__KY___HK______ 589 0.31%
_I_---S_________D___R_L____---G_---________________HTFPFANSNKSRNKAS___RYH__GY__KH___KY___HK______ 522 0.28%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR_L_KY___HK______ 507 0.27%
___---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 505 0.27%
_I_---S_________D_-________---E_---____________G___HTFP_-NSNKS_N_APS__RYH__GY__KH___KY___HK______ 477 0.25%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP_E_RYH_VGY__KR___KY___HK______ 441 0.23%
_I_---S__--_____D_-______-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 440 0.23%
_I_---S__--___________SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 438 0.23%
_I_---S_________D_-___L____---E_---___________V____HTFPFANSNKSRNKAPS__RYH__GY__KH_L_KY___HK______ 424 0.23%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__K-___KY___HK______ 418 0.22%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NRAPS__RYH__GYK_KH___KY___HK______ 408 0.22%
_I____S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNK_P___RYH_VGY__KR___KY___HK______ 333 0.18%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY_HKR___KY___HK______ 314 0.17%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK_____L 308 0.16%
_I_---S_V--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 280 0.15%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NIAPS__RYH__GY__KH___KY___HK______ 277 0.15%
_I_---S_________D_-________---E_YRR___________V____HTFPFANSNKS_NKAPS__RYH__GY__KH_L_KY___HK______ 251 0.13%
_I_---S_________D__________---E_---___________V____HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 245 0.13%
_I_---S_________D_-________---E_---___________V____HTFPFANS_KS_NKAPS__RYH__GY__KH___KY___HK______ 237 0.13%
_I_---S_________D_-________---E_---____________G___HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 236 0.13%
_I_---S__--_____D_-___S__-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 234 0.12%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KH___KY___HK______ 226 0.12%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NKAPS__RYH__GY_HKH___KY___HK______ 223 0.12%
_I_---S_________D_-________---E_---___________VG___HTFPFANSNKS_NKAPS__RYH__GY__KH___KY___HK______ 218 0.12%
_________--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 218 0.12%
_I_---S__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RY__VGY__KR___KY___HK______ 213 0.11%
_I_---S__--_____D_-___SG_-I---G_---_V______________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 198 0.11%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NRAPS__RYH__GY__KH_L_KY___HK______ 197 0.10%
_I__---__--_____D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 193 0.10%
_I_---S_________D_-________---E_---______N____V____HTFPFANSNKS_NRAPS__RYH__GY__KH___KY___HK______ 190 0.10%
_I_---S__--___I_D_-___SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 189 0.10%
_I__S___________D_-__G--__S---__---HV_--____________SFPFANSNKSRNKAP___RYH__GY__KH___KY___HK______ 178 0.09%
_I_---S__--_____D_____SG_-I---G_---________________H_FPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 173 0.09%
_I_---S_________D_-________---E_---___________V____HTFPFAN_NKS_NKAPS__RYH__GY__KH___KY___HK______ 159 0.08%
_I_---S_________D_-___L____---E_---___________V____HTFPFANSNKSWNKAPS__RYH__GY__KH___KY___HK______ 154 0.08%
_I_---S__--_____D_-___SG_-I---G_---________________HIFPFANSNKSWNKKP___RYH_VGY__KR___KY___HK______ 152 0.08%
_I_---S_________D_-________---E_---___________V____HTFPFANSNKS_NRAPS__RYHI_GY__KH___KY___HK______ 150 0.08%
Download
last modified: Wed May 24 14:07 2023
GISAID data provided on this website is subject to GISAID's Terms and Conditions
Questions or comments? Contact us at seq-info@lanl.gov.
Questions or comments? Contact us at seq-info@lanl.gov.




